The Human Breast Cell Atlas
comprehensively defining the cell types and states of the human breast.
Learn MoreThe Project
The anatomy and histopathology of the human breast has been studied for many decades and has provided insights into development, lactation and disease. More recently normal breast tissues have been profiled with molecular and genomic methods, which have largely focused on the epithelial cells. To date, an unbiased and comprehensive analysis of all cell types and their biological subtypes (cell states) has remained lacking. In this Human Breast Cell Atlas (HBCA) project, we aimed to generate a comprehensive reference of cell types and cell states in the adult human breast tissues using single cell and spatial genomic methods. Here, we profiled breast tissues from over 100 women with diverse biological states (parity, menopause, age, ethnicity), which has identified 12 major cell types and over 60 biological cell states. These cell types and cell states are spatially organized into four major areas of the breast: ductal, lobular, adipose and connective regions. Our analysis has revealed abundant pericyte, endothelial and immune cell populations in normal breast, as well as highly diverse luminal epithelial cell states. Our spatial mapping using multiple technologies revealed an unexpectedly rich ecosystem of tissue-resident immune cells in the ducts and lobules, as well as distinct epithelial cell state differences between ductal and lobular regions. Collectively, these data provide an unprecedented reference of adult normal breast tissue for studying mammary biology and disease states such as breast cancer. We are extremely grateful to all of the women who have selflessly donated their tissues to this project and provided this data to the research community. All of the single cell and spatial data from this project are available for open access via our data portals. This project has also established optimized tissue dissociation protocols and data analysis code that are freely available for download. Our project is part of the greater Human Cell Atlas (HCA) effort and has been generously funded by the Chan-Zuckerberg Initiative over many years. In addition to our HBCA project, there are several other ongoing human breast atlas projects that focus on different areas.
Learn MoreWorkflow Diagram
Overview and workflow of the HBCA project, in which breast tissue samples were collected from different sources, followed by single cell and single nucleus RNA sequencing (10x Genomics), as well as spatial analysis using CODEX (Akoya Biosciences), spatial transriptomics (Visium 10x Genomics), and smFISH (Resolve Biosciences and Vizgen Merscope).
220
Samples
Number of samples processed.
132
Women
The number of women participants.
714,331
Cells
Number of cells profiled using single cell RNA sequencing.
120,024
Nuclei
Number of nuclei profiled using single nucleus RNA sequencing.
Composition of the breast
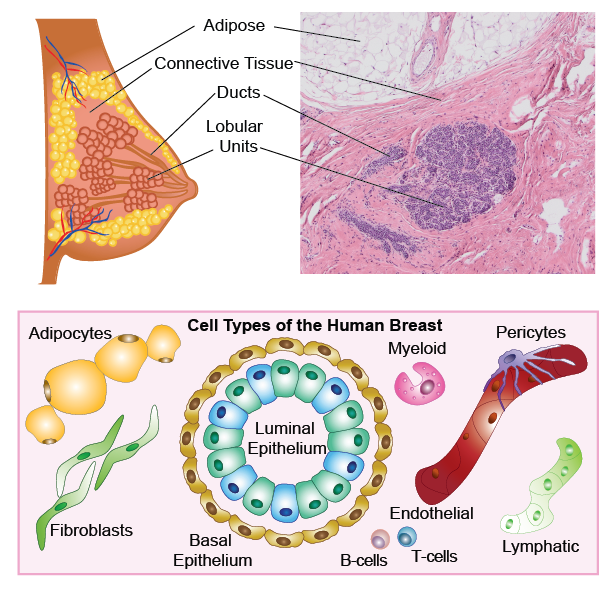
Anatomy of the adult human breast and pathological H&E section, with illustrations showing the major cell types.
Cell Types of the Breast
Dendogram of the different cell types as defined in the HBCA manuscript. Graphics Generated using BioRender.
Regional location of different cells types and states as determined using the spatial technologies in the manuscript. Graphics Generated using BioRender.
Single Cell Level UMAP
UMAP of scRNA-seq data from 714,331 cells integrated across over 100 tissues from over 150 women, showing 10 clusters that correspond to the major cell types. Note: 2 more clusters were noticed in the single nuclei data.
The Data Types
Single Cell Sequencing
10X Genomics Single Cell Chromium 3’ protocols from fresh cell suspesions.
Single Nuclei Sequencing
10X Genomics Single Cell Chromium 3’ protocols from nuceli suspesions.
Spatial Transcriptomics
10X Genomics Visium Spatial Gene Expression.
CODEX
Akoya Biosciences COdetection by inDEXing (CODEX®/PhenoCycler) 34-antibody panel.
smFISH
Segmentated Resolve Biosciences smFISH 100 gene panel.
smFISH
Segmentated Vizgen MERSCOPE custom 140-300 gene panels.
Our Team
MD Anderson Cancer Center
Nicholas E. Navin, Ph.D
Ken Chen, Ph.D
University of California, Irvine
Kai Kessenbrock, PhD
Devon Lawson, Ph.D
Baylor College of Medicine
Bora Lim, MD
Alastair Thompson, MD
Supported by
The HBCA was funded by the CZI and part of the Human Cell Atlas Network.